Semantic parsing of clinical protocols: a modest proposal
Clinical protocols
Clinical protocols are key to quality health care delivery. It may come as a surprise to some, but doctors and other clinicans don’t approach every patient as a creative blank slate: every hospital has numerous clinical protocols: well-defined instructions used to solve a class of specific problems. In other words, algorithms.
Now, as someone who works with algorithms in a somewhat professional capacity, I am used to algorithms being represented like this:
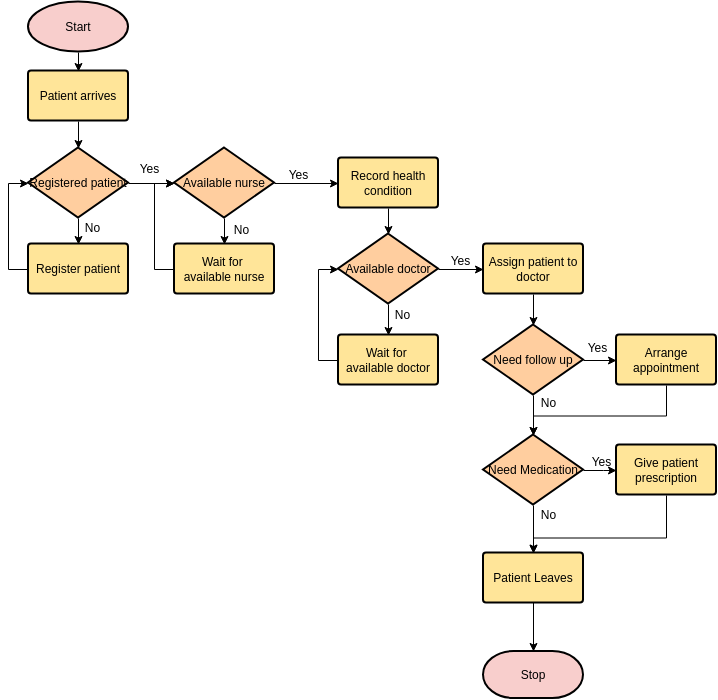
Or like this:
def patient_workflow(patient):
if not is_registered(patient):
register(patient)
nurse = wait_for_available_nurse()
record_health_condition(patient, nurse)
doctor = wait_for_available_doctor()
assign_doctor(patient, doctor)
if follow_up_required(patient):
appointment = arrange_appointment(patient, doctor)
if medication_required(patient):
prescribe_medication(patient, doctor)
Or, on a really good day, like this:
(def nurse-ref (delay (wait-for-available-nurse)))
(def doctor-ref (delay (wait-for-available-doctor)))
(defn apply-if [cond f]
(fn [arg] (if (cond arg) (f arg) arg)))
(defn rcomp [& args]
(apply comp (reverse args))
(def steps [
(apply-if (complement registered?) register)
(fn [patient] (record-health-condition patient @nurse))
(fn [patient] (assign-doctor patient @doctor))
(apply-if follow-up-required? arrange-appointment)
(apply-if medication-required? prescribe-medication)])
(def patient-workflow (rcomp steps))
But definitley not as 81-page PDFs in plain English. Which is how most clinical protocols tend to look.
So what?
There are a number of advantages to representing algorithms in a language that computers can easily understand. The main ones are
- Static analysis. As a poignant observation by Nathaniel Benchley goes, “anyone can do any amount of work, provided it isn’t the work he is supposed to be doing at that moment”. For programmers this often takes form of, instead of writing the software their clients directly needs, developing static analysis tools that help them (maybe, in the future) develop software their clients need. These tools can find security vulnerabilities, enforce code style guidelines, detect code that never gets executed (“dead code”), highlight common mistakes and inefficiencies and more. The term of the trade for this is Computer-Aided Software Engineering. These tools can detect potential inefficiencies in hospital workflows just as well as mobile apps, provided that hospital workflows are described in a language the tool was built for.
- Automation. Some steps of the clinical alg… sorry, protocols, can be automated with existing technology. See “arrange appointment” in the example above - this is something your phone does just as well as most appoinment-arranging professionals. Other steps will become automatable in the future. By expressing clinical protocols in a programming language, we are laying the groundwork for clinical decision support systems of the future.
- Simulation. Patient simulator software can be used to test clinical protocols in a virtual environment. New protocols can be tested in a simulator before going through a long and expensive clinical trial. Morevoer, modern genetic programming techniques can be used to not only test protocols in a simulator, but generate better protocols automatically. This is the end goal of deep program inducation for personal healthcare research.
But how?
The task of converting a natural language description of an algorithm into programming language is known as semantic parsing [Lee te al 2021]. It is a specific case of sequence to sequence problem and, as such we should expect seq2seq machine learning models (mainly recurrent neural network-based encoder decoder models) to work, however, programming languages present a number of additional challenges. See Autoencoders as Tools for Program Synthesis [de Bruin et al 2021], NetSyn [Mandal et al 2019], AIProgrammer [Becker, Gottschlich 2017] and A survey of Machine Learning for Big Code and Naturalness as well as this ever-updating list on github
An important skill for people wokring in these field is dataset hacking: the web is full of textual descriptions of algorithms and even more full of code, however, the perfect dataset for semantic parsing consists of pairs of strings describing the same algorithm in natural and machine language. One can tackle this issue with unsupervised learning [Artetxe et al 2017] or come up with clever ways to find semantic parsing datasets in unobvious places. Textbooks that give a natural language introduction of an algorithm, followed by pseudocode, followed by actual executable code. Research papers with code artefacts attached to them. Or… collectible card games! Many games started out as tabletop card games where every card has a description of what happens once it’s played written on the front of the card and then were turned into computer games. The result? A dataset of these

The state of the art paper on semantic parsing uses this dataset to train an LSTM-based encoder with a tree-based decoder:

That said, the field is quite new and which methods turn out to be the best remains to be seen. New methods will inevitably arise from further research.
Confessions
This work is taxpayer funded